First overview of a measurement
Measuring NMR spectra of samples often results in a series of measurements, and one of the first tasks is to get an overview what has been measured and how the results look like.
Classes used:
Processing
Plotting
Recipe
Complete recipe for getting an overview and compare two spectra.
1format:
2 type: ASpecD recipe
3 version: '0.2'
4settings:
5 default_package: nmraspecds
6directories:
7 datasets_source: ../../tests/testdata/
8datasets:
9- source: Adamantane/1
10 id: 1H
11 importer_parameter:
12 processing_number: 2
13- source: Adamantane/2
14 id: 13C
15tasks:
16 - kind: singleplot
17 type: SinglePlotter1D
18 properties:
19 properties:
20 figure:
21 dpi: 600
22 filename:
23 - ProtonSpectrum.png
24 - CarbonSpectrum.png
25 - kind: processing
26 type: Normalisation
27 - kind: multiplot
28 type: MultiPlotter1D
29 properties:
30 properties:
31 figure:
32 dpi: 600
33 axes:
34 xlim: [100, -100]
35 parameters:
36 offset: 1
37 filename: BothSpectra.png
38 apply_to:
39 - 13C
40 - 1H
Result
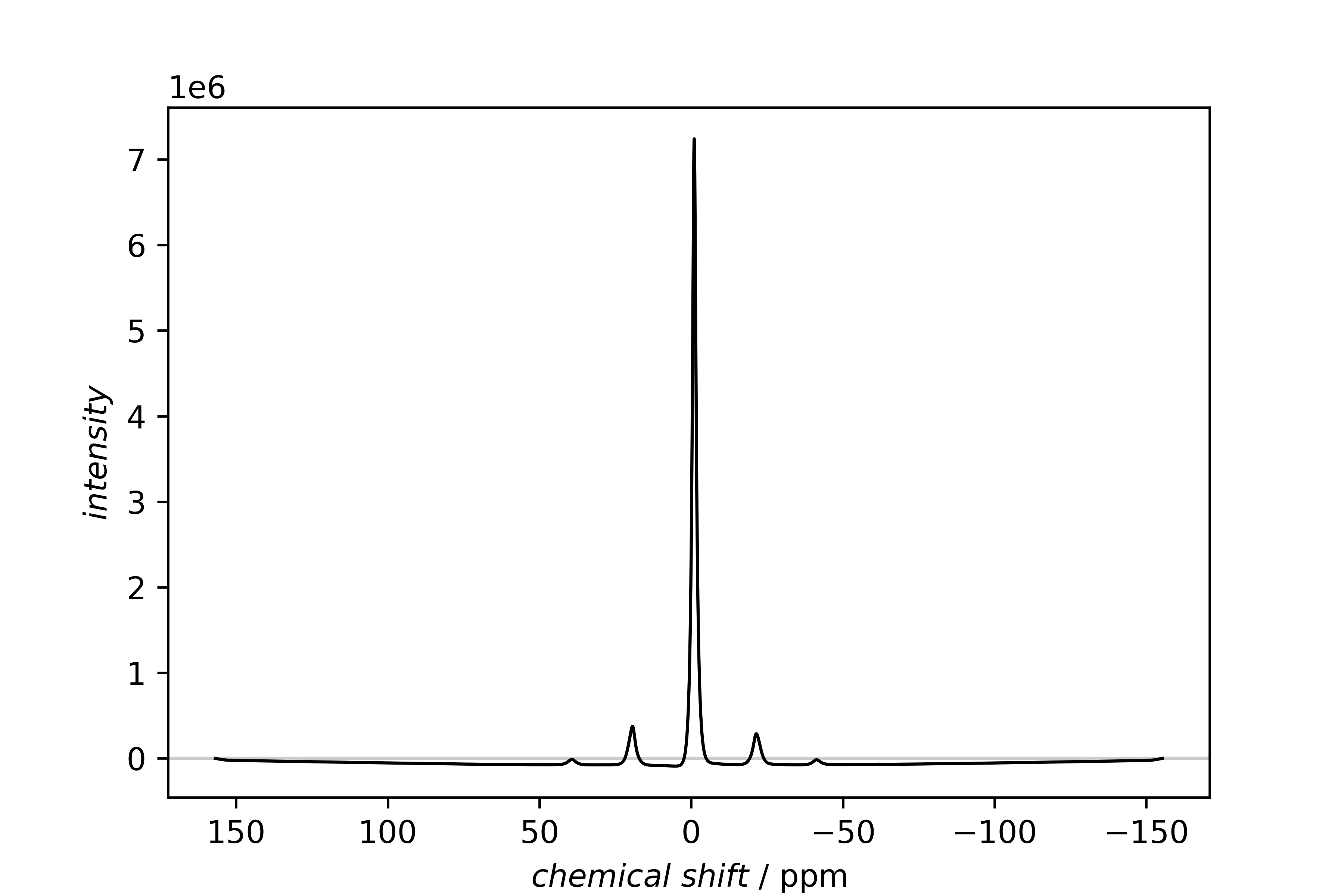
1H spectrum
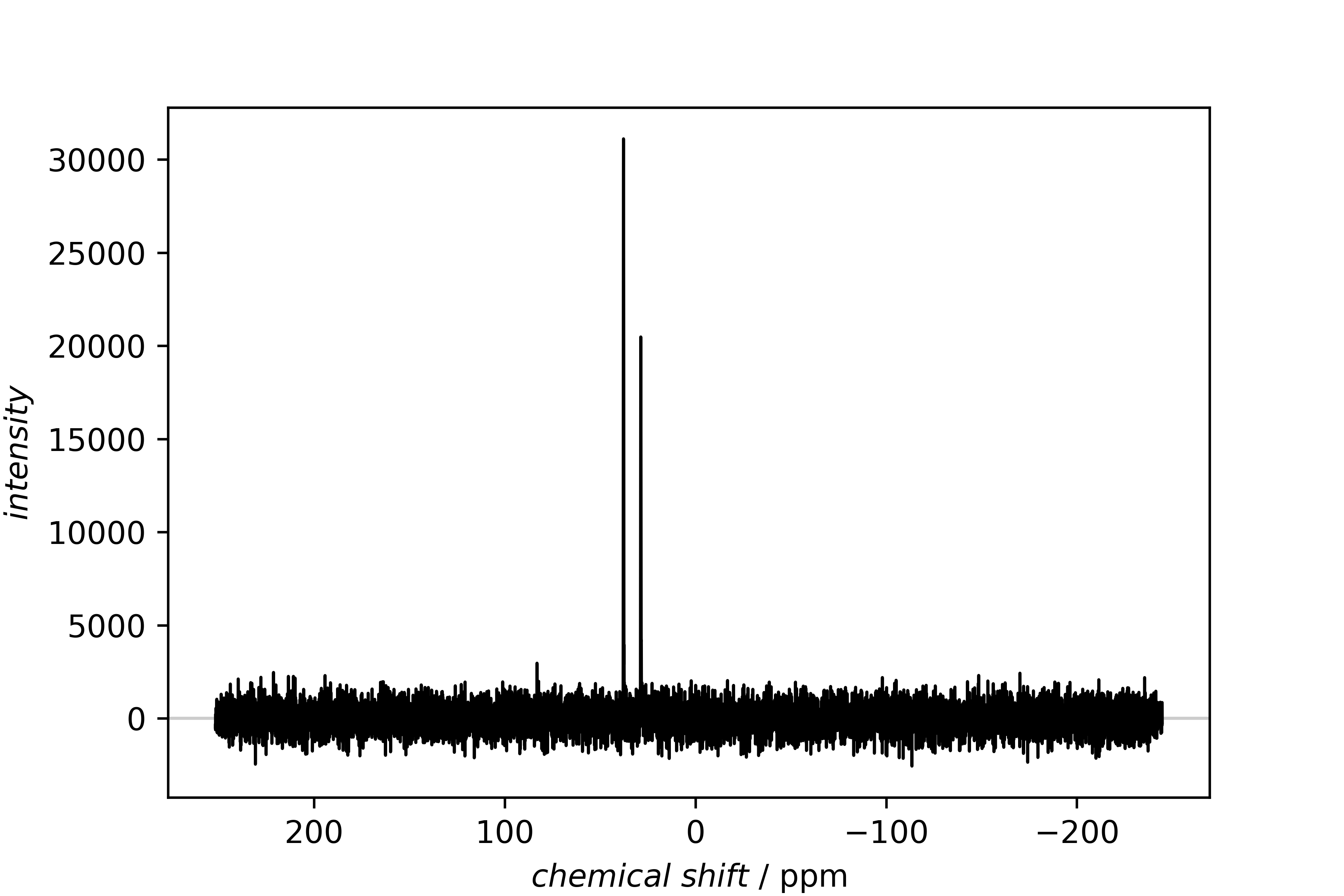
13C spectrum
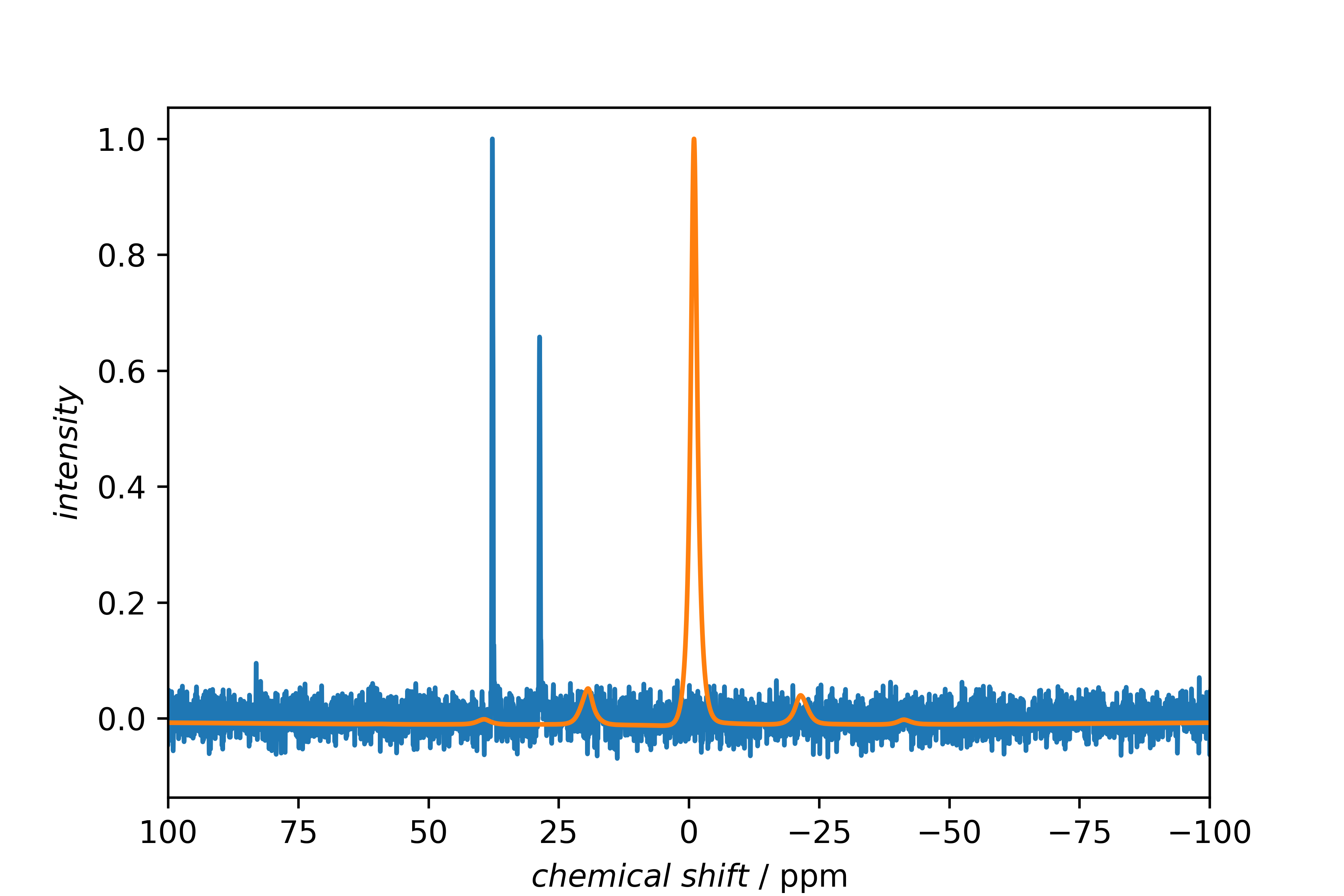
Both spectra in one figure.
Comments
The recipe starts with information about its version, the default package and the source path of the datasets.
At the import, the second processing number is called for the 1H dataset, indicated by the importer_parameter keyword.
The 13C data is imported without any further parameters, meaning, that the first processing number is imported
Both datasets get an ID with which they are called later.
Directly after the import, both spectra are plotted separately with the SinglePlotter1D.
Then, they are normalized, without further parameters, this is done using the maximum.
Both spectra are plotted into one figure (ignoring that plotting a 1H spectrum over a 13C spectrum physically makes nearly no sense).